In insects, wings are subjected to strong natural selection pressures for camouflage, mimicry and warning coloration. The patterns and colours of butterfly wings, for example, are good models for studying these processes. Changes at the genetic level cause changes in the combination of pigments and their relative position during larval wing development and metamorphosis.
In Latin and South America, butterflies of the genus Heliconius are particularly interesting on this subject. They have a wide pattern diversity (pattern type) but converge in each geographic region to the point that all species are similar. For many years, scientists have been trying to understand the genetic and evolutionary mechanisms behind this morphological diversity.
The team, led by Richard W.R. Wallbank (University of Cambridge), provides some answers. Through complex genetic analyses of several species of Heliconius, these scientists have highlighted the recombination and introgression of certain genes that have occurred between species and populations during their evolutionary history. These mechanisms favour the emergence of new patterns which will be selected in populations and which will thus allow species to evolve rapidly.
One of the main obstacles to evolutionary innovation is the constraint of genetic change imposed by the existing function. Indeed, mutations conferring advantageous phenotypes (appearance of a morphological trait) for a novel trait often cause negative effects on other traits influenced by the gene concerned ( pleiotropic effect).
This pleiotropic functioning of certain genes restricts the appearance of phenotypic novelties and thus the diversity of species. For example, genes controlling wing development in insects have been altered little during evolution because they are subject to high selection pressure. The expression profiles of these genes are thus similar between flies and butterflies.
caption id=”attachment_2823″ align=”alignright” width=”339″] Figure 1 : Diversity of patterns of several Amazonian species – 1st line : H. burneyi huebneri, H. aoede auca and H. xanthocles Zamora – 2nd line : H. timareta f. timareta, H. doris doris and H. demeter ucayalensis – 3rd line : H. melpomene malleti, H. egeria homogena and H. erato emma – 4th line : H. elevatus pseudocupidineus, Eueides heliconioides eanes and E. tales calathus – 5th line : Cretonne phyllies (Source : Wallbank et al, 2016)[/caption]
Figure 1 : Diversity of patterns of several Amazonian species – 1st line : H. burneyi huebneri, H. aoede auca and H. xanthocles Zamora – 2nd line : H. timareta f. timareta, H. doris doris and H. demeter ucayalensis – 3rd line : H. melpomene malleti, H. egeria homogena and H. erato emma – 4th line : H. elevatus pseudocupidineus, Eueides heliconioides eanes and E. tales calathus – 5th line : Cretonne phyllies (Source : Wallbank et al, 2016)[/caption]
However, studies have shown that genetic structures exist to circumvent these constraints. They are made up of juxtaposed elements, called modules, coding for specific domains, where the modification of one of them does not disturb the expression of the others. This principle seems to be at the origin of morphological diversity.
Such elements have been discovered in the Heliconius (thematic I worked on in 2009, see at the bottom of the page). These butterflies, which are found in Latin and South America, gather many species with patterns and colors of great diversity. In the same geographical locality, all the species have the same wing colour structure. This mimetic convergence is particularly remarkable for the 11 species of Heliconius with the phenotype called “Dennis-Ray”: red spots at the base of the forewings (Dennis) and red stripes on hindwings (Ray) (see figure 1).
What are the genetic and evolutionary processes that have allowed all these species, sometimes phylogenetically distant, to evolve towards a similar pattern?
This is what the Wallbank team wanted to answer.
For this, scientists have undertaken genetic analyses on Heliconius melpomene and related species such as H. elevatus and H. timareta to identify the modules associated with the elements at the origin of the phenotypes Dennis and Ray.
Their results reveal a more complex evolutionary history than scientists assumed !
Genomic analyses were performed from 96 specimens belonging to a population of H. melpomene from French Guiana : H. m. meriana which does not show scratches on the hindwings; to a population of H. timareta from Ecuador : H. t. timareta forme contigua not having Dennis patterns on the forewings H hybrid individuals. melpomene from Ecuador with no scratches on the hindwings ( see figure 2).
caption id=”attachment_2827″ align=”aligncenter” width=”435″] Figure 2 : Examples of the main Dennis-Ray patterns of the study – H. e. pseudocupidineus and H. m. malleti (Dennis-Ray), H. m. meriana (Dennis), H. t. timareta f. contitigua (Ray) and H. m. rosina (other) (Source : Wallbank et al., 2016)[/caption]
Figure 2 : Examples of the main Dennis-Ray patterns of the study – H. e. pseudocupidineus and H. m. malleti (Dennis-Ray), H. m. meriana (Dennis), H. t. timareta f. contitigua (Ray) and H. m. rosina (other) (Source : Wallbank et al., 2016)[/caption]
After these analyses, scientists discovered two distinct regions, each sequence being associated with one of the two phenotypes (10 000 and 25 000 base pairs for the Dennis and Ray phenotypes respectively) ( see figure 3).
caption id=”attachment_2830″ align=”aligncenter” width=”471″] Figure 3 : Genome regions associated with Dennis and Ray phenotypes – D/d : Dennis expressed/absent – R/r : Ray expressed/absent (Source : Wallbank et al., 2016)[/caption] Then, using a method called Base Local Alignment Search Tool (BLAST), scientists aligned different sequences of these individuals to compare them with each other but also with that of H. cydno, not having stripes and that of a reference species, more distant phylogenetically and belonging to the clade of Silvaniformes : H. elevatus.
Figure 3 : Genome regions associated with Dennis and Ray phenotypes – D/d : Dennis expressed/absent – R/r : Ray expressed/absent (Source : Wallbank et al., 2016)[/caption] Then, using a method called Base Local Alignment Search Tool (BLAST), scientists aligned different sequences of these individuals to compare them with each other but also with that of H. cydno, not having stripes and that of a reference species, more distant phylogenetically and belonging to the clade of Silvaniformes : H. elevatus.
The results indicate that the Dennis and Ray phenotypes are controlled by distinct adjacent genetic elements, where each of them expresses itself differently within each species and subpopulation of Heliconius, thus giving them a specific pattern.
Scientists have been able to reconstruct the evolutionary history of these genetic elements within these species using this news on the genetic architecture of the alleles (version of a gene) at the origin of the Dennis and Ray phenotypes. This is based on multiple successive and recent phenomena of introgression (gene exchange between species) and recombination between the three species: H. elevatus, H. melpomene and H. timareta ( see figure 4).
caption id=”attachment_2833″ align=”alignright” width=”360″] Figure 4 : Hypothesis of the origin of the introgressions of the Dennis and Ray alleles between the species H. elevatus, H. melpomene and H. timareta (Source : Wallbank et al., 2016)[/caption]
Figure 4 : Hypothesis of the origin of the introgressions of the Dennis and Ray alleles between the species H. elevatus, H. melpomene and H. timareta (Source : Wallbank et al., 2016)[/caption]
Dennis allele currently present in H. melpomene would come from an ancestral form of H. elevatus, a transmission that occurred 2 million years ago, 2 million years after these species diverged. The species of the Silvaniforme clade mimic butterflies of the Ithomiini tribe and are marbled, orange/red in appearance with black and yellow “tiger” patterns. The orange spots at the base of their forewings, similar to the Dennis phenotypes of H. melpomene, seem to confirm this ancestral origin.
A second introgression event of this allele would have occurred more recently, 0.45 million years ago, around H. timareta.
The allele to the Ray phenotype organ would have been transmitted a first time from H. melpomene to H. timareta a little over a million years ago, then a second time around H. elevatus 400,000 years later.
So, during evolution, the wing patterns of these species change. For example, the Dennis form was the only one represented in Guyana populations for 1 million years and then evolved to the current Dennis-Ray form.
Low levels of genetic diversity between these alleles within species confirm the recent origin of these introgressions : alleles differ by less than 2% (molecular clock : hypothesis that genetic mutations accumulate in a genome at a rate globally proportional to geological time).
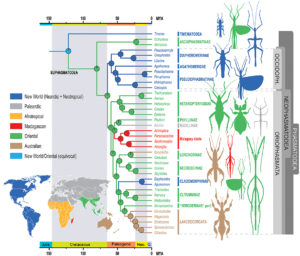
Comments: a form of H. melpomene, H. m. meriana, no longer has the Ray phenotype. The loss of this phenotype results from recombination at this allele preventing it from expressing itself. The same event occurred for the Dennis and Ray alleles in H. t. timareta, almost entirely black ( see figure 5).
This study brings new elements to better understand how evolution works and highlights the genetic mechanisms at work leading to the appearance of new morphological forms. Introgressions and recombinations seem to explain the evolutionary history of other species such as Darwin’s finches, sticklebacks or even… man.
caption id=”attachment_2838″ align=”aligncenter” width=”500″] Figure 5 : Phylogeny of the species studied and their respective combinations in Dennis and Ray alleles (Source : Wallbank et al., 2016)[/caption]
Figure 5 : Phylogeny of the species studied and their respective combinations in Dennis and Ray alleles (Source : Wallbank et al., 2016)[/caption]
Source :
– Wallbank R.W.R. et al. (2016) : Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 14(1):1-16 (lien)
Photo and video albums of Heliconius
Photos and videos taken by Benoît GILLES during his stays at Panama and French Guiana in the framework of studies on Heliconius at Muséum National d’Histoire Naturelle de Paris : research works that have been the subject of a publication to be read here